
CEIT-UGA
A Data Analytics Framework for Mining the Dark Kinome
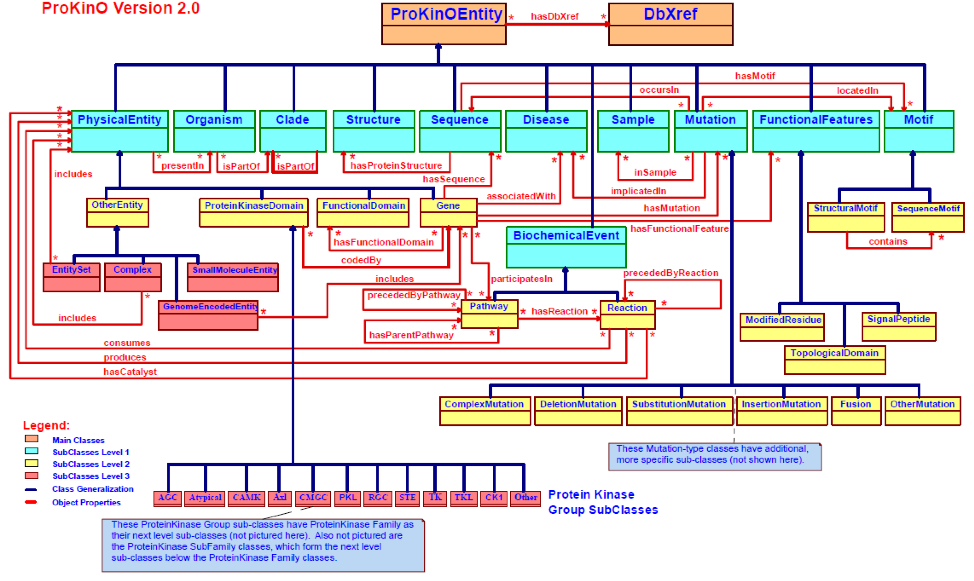
ProKinO: https://vulcan.cs.uga.edu/prokino/about/prokino

Natarajan Kannan, Ph.D.
Associate Professor
Biochemistry and Molecular Biology
Associate Director of Institute of Bioinformatics
University of Georgia

Krzysztof J. Kochut, Ph.D.
Professor
Department of Computer Science
University of Georgia
Contacts
Lab websites:
- Natarajan Kannan profile: https://www.bmb.uga.edu/directory/people/natarajan-kannan
- Krzysztof J. Kochut profile: https://cobweb.cs.uga.edu/~kochut/
Overview
Collaborative work by Dr. Natarajan Kannan and Dr. Krzysztof Kochut have resulted in the development of Protein Kinase Ontology (ProKinO) for organizing information about kinase structure, function and evolution. The aim is to further this work by developing a novel comparative kinomics framework applicable to visualizing the understudied kinases in these organizational context. And build an interface for interacting with this framework and queries for hypothesis generation and knowledge discovery.
NIH grant number: 1U01 CA239106
Publications:
- O'Boyle B, Shrestha S, Kochut K, Eyers PA, Kannan N. Computational tools and resources for pseudokinase research. Methods Enzymol. 2022;667:403-426. doi: 10.1016/bs.mie.2022.03.040. Epub 2022 Apr 8. PMID: 35525549; PMCID: PMC9733567.
- Harris JA, Fairweather E, Byrne DP, Eyers PA. Analysis of human Tribbles 2 (TRIB2) pseudokinase. Methods Enzymol. 2022;667:79-99. doi: 10.1016/bs.mie.2022.03.025. Epub 2022 Apr 13. PMID: 35525562.
- Shrestha S, Bendzunas G, Kannan N. Protein kinase inhibitor selectivity "hinges" on evolution. Structure. 2022 Dec 1;30(12):1561-1563. doi: 10.1016/j.str.2022.11.004. PMID: 36459973.
- Yeung W, Ruan Z, Kannan N. Emerging roles of the αC-β4 loop in protein kinase structure, function, evolution, and disease. IUBMB Life. 2020 Jun;72(6):1189-1202. doi: 10.1002/iub.2253. Epub 2020 Feb 26. PMID: 32101380; PMCID: PMC8375298.
- Shrestha S, Byrne DP, Harris JA, Kannan N, Eyers PA. Cataloguing the dead: breathing new life into pseudokinase research. FEBS J. 2020 Oct;287(19):4150-4169. doi: 10.1111/febs.15246. Epub 2020 Mar 10. PMID: 32053275; PMCID: PMC7586955.
- Cao M, Day AM, Galler M, Latimer HR, Byrne DP, Foy TW, Dwyer E, Bennett E, Palmer J, Morgan BA, Eyers PA, Veal EA. A peroxiredoxin-P38 MAPK scaffold increases MAPK activity by MAP3K-independent mechanisms. Mol Cell. 2023 Sep 7;83(17):3140-3154.e7. doi: 10.1016/j.molcel.2023.07.018. Epub 2023 Aug 11. PMID: 37572670.
- Soleymani S, Gravel N, Huang LC, Yeung W, Bozorgi E, Bendzunas NG, Kochut KJ, Kannan N. Dark kinase annotation, mining, and visualization using the Protein Kinase Ontology. PeerJ. 2023 Dec 5;11:e16087. doi: 10.7717/peerj.16087. PMID: 38077442; PMCID: PMC10704995.
- Huang LC, Yeung W, Wang Y, Cheng H, Venkat A, Li S, Ma P, Rasheed K, Kannan N. Quantitative Structure-Mutation-Activity Relationship Tests (QSMART) model for protein kinase inhibitor response prediction. BMC Bioinformatics. 2020 Nov 12;21(1):520. doi: 10.1186/s12859-020-03842-6. PMID: 33183223; PMCID: PMC7664030.
- Byrne DP, Shrestha S, Daly LA, Marensi V, Ramakrishnan K, Eyers CE, Kannan N, Eyers PA. Evolutionary and cellular analysis of the 'dark' pseudokinase PSKH2. Biochem J. 2023 Jan 31;480(2):141-160. doi: 10.1042/BCJ20220474. PMID: 36520605; PMCID: PMC9988210.
- Anandakrishnan M, Ross KE, Chen C, Shanker V, Cowart J, Wu CH. KSFinder-a knowledge graph model for link prediction of novel phosphorylated substrates of kinases. PeerJ. 2023 Oct 6;11:e16164. doi: 10.7717/peerj.16164. PMID: 37818330; PMCID: PMC10561642.
- PDBe-KB consortium. PDBe-KB: a community-driven resource for structural and functional annotations. Nucleic Acids Res. 2020 Jan 8;48(D1):D344-D353. doi: 10.1093/nar/gkz853. PMID: 31584092; PMCID: PMC6943075.
- Salcedo MV, Gravel N, Keshavarzi A, Huang LC, Kochut KJ, Kannan N. Predicting protein and pathway associations for understudied dark kinases using pattern-constrained knowledge graph embedding. PeerJ. 2023 Oct 18;11:e15815. doi: 10.7717/peerj.15815. PMID: 37868056; PMCID: PMC10590106.
- Agnew C, Liu L, Liu S, Xu W, You L, Yeung W, Kannan N, Jablons D, Jura N. The crystal structure of the protein kinase HIPK2 reveals a unique architecture of its CMGC-insert region. J Biol Chem. 2019 Sep 13;294(37):13545-13559. doi: 10.1074/jbc.RA119.009725. Epub 2019 Jul 24. PMID: 31341017; PMCID: PMC6746438.
- Baffi TR, Lordén G, Wozniak JM, Feichtner A, Yeung W, Kornev AP, King CC, Del Rio JC, Limaye AJ, Bogomolovas J, Gould CM, Chen J, Kennedy EJ, Kannan N, Gonzalez DJ, Stefan E, Taylor SS, Newton AC. mTORC2 controls the activity of PKC and Akt by phosphorylating a conserved TOR interaction motif. Sci Signal. 2021 Apr 13;14(678):eabe4509. doi: 10.1126/scisignal.abe4509. PMID: 33850054; PMCID: PMC8208635.
- Huang LC, Taujale R, Gravel N, Venkat A, Yeung W, Byrne DP, Eyers PA, Kannan N. KinOrtho: a method for mapping human kinase orthologs across the tree of life and illuminating understudied kinases. BMC Bioinformatics. 2021 Sep 18;22(1):446. doi: 10.1186/s12859-021-04358-3. PMID: 34537014; PMCID: PMC8449880.
- Picado A, Chaikuad A, Wells CI, Shrestha S, Zuercher WJ, Pickett JE, Kwarcinski FE, Sinha P, de Silva CS, Zutshi R, Liu S, Kannan N, Knapp S, Drewry DH, Willson TM. A Chemical Probe for Dark Kinase STK17B Derives Its Potency and High Selectivity through a Unique P-Loop Conformation. J Med Chem. 2020 Dec 10;63(23):14626-14646. doi: 10.1021/acs.jmedchem.0c01174. Epub 2020 Nov 20. PMID: 33215924; PMCID: PMC7816213.
