
Knowledge Management Center for Illuminating the Druggable Genome

Jeremy Edwards, Ph.D
Professor and Department Chairman
Department of Chemistry and Chemical Biology
University of New Mexico Translational Informatics Division
Department of Internal Medicine
Health Sciences Center
University of New Mexico
Previous PI (Jan. 2018 - Jan. 2022)

Tudor Oprea, Ph.D., M.D. Professor
Chief, Translational Informatics Division
Department of Internal Medicine
Health Sciences Center
University of New Mexico
Contacts
- Translational Informatics Division: https://datascience.unm.edu/
- TCRD (Target Central Resource Database): http://juniper.health.unm.edu/tcrd/
- REST API for TCRD (underlying data for Pharos): http://juniper.health.unm.edu/tcrd/api.html
- TINx (Target Importance and Novelty eXplorer): https://newdrugtargets.org/
- DrugCentral (utilized to annotate Tclin): https://drugcentral.org/
![]() Translational Informatics: @translationalID
Translational Informatics: @translationalID
Overview
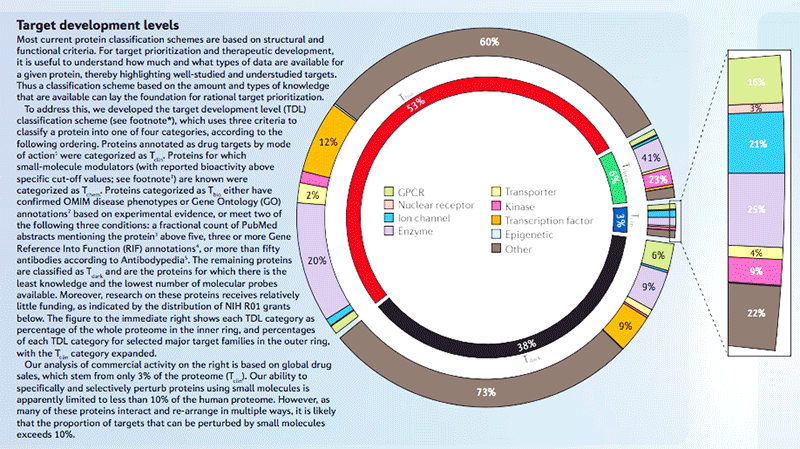
KMC grant at UNM is led by Dr. Jeremy Edwards and includes collaborations with Dr. Lars Juhl Jensen’s lab at NNFCPR, EMBL-EBI with Dr. Andrew Leach as their Group Lead, and Dr. Soren Avrim at ICTRA. Previous PI for the KMC grant at UNM was Dr. Tudor Oprea. Work from UNM includes the development of TCRD, the main database supporting Pharos, and appropriate visualization tools, such as TINx. Work by Dr. Oprea’s group include evaluation of the data collated into TCRD and led to the categorization of target proteins based on knowledge levels, Target Development Levels (TDL): Tdark are targets with least knowledge. Next level of development are Tbio, proteins with further annotation of function or biological process, including confirmed OMIM phenotypes. With the discovery of protein interacting with a drug or compound, these proteins are classified as Tchem if these compound activities satisfy activity thresholds set based on protein biological class. Further annotation via DrugCentral regarding the mechanism of action for approved drugs elevates the target into the TDL of Tclin, i.e., protein with highest attained annotation.
NIH grant number: 1U24 CA224370-01
Nature Rev. Drug Discov. Poster:
Unexplored opportunities in the druggable human genome, T. Oprea, et al., Nature Rev. Drug Discov. Poster Jan. 2017.
KMC-UNM Publications:
- Overview of the Knowledge Management Center for Illuminating the Druggable Genome.
Oprea TI, Bologa C, Holmes J, Mathias S, Metzger VT, Waller A, Yang JJ, Leach AR, Jensen LJ, Kelleher KJ, Sheils TK, Mathé E, Avram S, Edwards JS. Drug Discov Today. 2024 Mar;29(3):103882. doi: 10.1016/j.drudis.2024.103882. Epub 2024 Jan 11. PMID: 38218214 - Exploring DrugCentral: from molecular structures to clinical effects.
Halip L, Avram S, Curpan R, Borota A, Bora A, Bologa C, Oprea TI. J Comput Aided Mol Des. 2023 Dec;37(12):681-694. doi: 10.1007/s10822-023-00529-x. Epub 2023 Sep 14. Erratum in: J Comput Aided Mol Des. 2023 Dec 2;38(1):2. PMID: 37707619; PMCID:PMC10692006. - Molecular Complexity: You Know It When You See It.
Oprea TI, Bologa C. J Med Chem. 2023 Sep 28;66(18):12710-12714. doi: 10.1021/acs.jmedchem.3c01507. Epub 2023 Sep 7. PMID: 37675804; PMCID: PMC10544322. - Pharos 2023: an integrated resource for the understudied human proteome.
Kelleher KJ, Sheils TK, Mathias SL, Yang JJ, Metzger VT, Siramshetty VB, Nguyen DT, Jensen LJ, Vidović D, Schürer SC, Holmes J, Sharma KR, Pillai A, Bologa CG, Edwards JS, Mathé EA, Oprea TI. Nucleic Acids Res. 2023 Jan 6;51(D1):D1405-D1416. doi: 10.1093/nar/gkac1033. Erratum in: Nucleic Acids Res. 2023 Feb 28;51(4):1999. PMID: 36624666; PMCID: PMC9825581. - DrugCentral 2023 extends human clinical data and integrates veterinary drugs.
Avram S, Wilson TB, Curpan R, Halip L, Borota A, Bora A, Bologa CG, Holmes J, Knockel J, Yang JJ, Oprea TI. Nucleic Acids Res. 2023 Jan 6;51(D1):D1276-D1287. doi: 10.1093/nar/gkac1085. PMID: 36484092; PMCID: PMC9825566. - CACHE (Critical Assessment of Computational Hit-finding Experiments): A public-private partnership benchmarking initiative to enable the development of computational methods for hit-finding.
Ackloo S, Al-Awar R, Amaro RE, Arrowsmith CH, Azevedo H, Batey RA, Bengio Y, Betz UAK, Bologa CG, Chodera JD, Cornell WD, Dunham I, Ecker GF, Edfeldt K, Edwards AM, Gilson MK, Gordijo CR, Hessler G, Hillisch A, Hogner A, Irwin JJ, Jansen JM, Kuhn D, Leach AR, Lee AA, Lessel U, Morgan MR, Moult J, Muegge I, Oprea TI, Perry BG, Riley P, Rousseaux SAL, Saikatendu KS, Santhakumar V, Schapira M, Scholten C, Todd MH, Vedadi M, Volkamer A, Willson TM. Nat Rev Chem. 2022 Apr;6(4):287-295. doi: 10.1038/s41570-022-00363-z. Epub 2022 Feb 15. PMID: 35783295; PMCID: PMC9246350. - Autophagy and Herpesvirus: A collaboration Contributing to Alzheimer's Disease.
Nafchi AR, Esmaeili M, Myers O, Oprea T, Bearer EL. FASEB J. 2022 May;36 Suppl 1. doi: 10.1096/fasebj.2022.36.S1.R2731. PMID: 35723876. - AlphaFold illuminates half of the dark human proteins.
Binder JL, Berendzen J, Stevens AO, He Y, Wang J, Dokholyan NV, Oprea TI. Curr Opin Struct Biol. 2022 Jun;74:102372. doi: 10.1016/j.sbi.2022.102372. Epub 2022 Apr 16. PMID: 35439658; PMCID: PMC10669925. - State of the Art and Uses for the Biopharmaceutics Drug Disposition Classification System (BDDCS): New Additions, Revisions, and Citation References.
Bocci G, Oprea TI, Benet LZ. AAPS J. 2022 Feb 23;24(2):37. doi: 10.1208/s12248-022-00687-0. PMID: 35199251; PMCID: PMC8865883. - Machine learning prediction and tau-based screening identifies potential Alzheimer's disease genes relevant to immunity.
Binder J, Ursu O, Bologa C, Jiang S, Maphis N, Dadras S, Chisholm D, Weick J, Myers O, Kumar P, Yang JJ, Bhaskar K, Oprea TI. Commun Biol. 2022 Feb 11;5(1):125. doi: 10.1038/s42003-022-03068-7. PMID: 35149761; PMCID: PMC8837797. - Getting Started with the IDG KMC Datasets and Tools.
Kropiwnicki E, Binder JL, Yang JJ, Holmes J, Lachmann A, Clarke DJB, Sheils T, Kelleher KJ, Metzger VT, Bologa CG, Oprea TI, Ma'ayan A. Curr Protoc. 2022 Jan;2(1):e355. doi: 10.1002/cpz1.355. PMID: 35085427; PMCID: PMC10789444. - Supervised learning with word embeddings derived from PubMed captures latent knowledge about protein kinases and cancer.
Ravanmehr V, Blau H, Cappelletti L, Fontana T, Carmody L, Coleman B, George J, Reese J, Joachimiak M, Bocci G, Hansen P, Bult C, Rueter J, Casiraghi E, Valentini G, Mungall C, Oprea TI, Robinson PN. NAR Genom Bioinform. 2021 Dec 8;3(4):lqab113. doi: 10.1093/nargab/lqab113. PMID: 34888523; PMCID: PMC8652379. - A critical overview of computational approaches employed for COVID-19 drug discovery.
Muratov EN , Amaro R , Andrade CH , Brown N , Ekins S , Fourches D , Isayev O , Kozakov D , Medina-Franco JL , Merz KM , Oprea TI , Poroikov V , Schneider G , Todd MH , Varnek A , Winkler DA , Zakharov AV , Cherkasov A , Tropsha A . Chem Soc Rev. 2021 Aug 21;50(16):9121-9151. doi: 10.1039/d0cs01065k. Epub 2021 Jul 2. PMID: 34212944; PMCID: PMC8371861. - TIGA: target illumination GWAS analytics.
Yang JJ, Grissa D, Lambert CG, Bologa CG, Mathias SL, Waller A, Wild DJ, Jensen LJ, Oprea TI. Bioinformatics. 2021 Nov 5;37(21):3865-3873. doi: 10.1093/bioinformatics/btab427. PMID: 34086846. - Crowdsourced mapping of unexplored target space of kinase inhibitors.
Cichońska A, Ravikumar B, Allaway RJ, Wan F, Park S, Isayev O, Li S, Mason M, Lamb A, Tanoli Z, Jeon M, Kim S, Popova M, Capuzzi S, Zeng J, Dang K, Koytiger G, Kang J, Wells CI, Willson TM; IDG-DREAM Drug-Kinase Binding Prediction Challenge Consortium; Oprea TI, Schlessinger A, Drewry DH, Stolovitzky G, Wennerberg K, Guinney J, Aittokallio T. Nat Commun. 2021 Jun 3;12(1):3307. doi: 10.1038/s41467-021-23165-1. PMID: 34083538; PMCID: PMC8175708. - Highly Accurate Chip-Based Resequencing of SARS-CoV-2 Clinical Samples.
Hoff K, Ding X, Carter L, Duque J, Lin JY, Dung S, Singh P, Sun J, Crnogorac F, Swaminathan R, Alden EN, Zhu X, Shimada R, Posavi M, Hull N, Dinwiddie D, Halasz AM, McGall G, Zhou W, Edwards JS. Langmuir. 2021 Apr 27;37(16):4763-4771. doi: 10.1021/acs.langmuir.0c02927. Epub 2021 Apr 13. PMID: 33848173; PMCID: PMC8056606. - InContext: curation of medical context for drug indications.
Moodley K, Rieswijk L, Oprea TI, Dumontier M. J Biomed Semantics. 2021 Feb 12;12(1):2. doi: 10.1186/s13326-021-00234-4. PMID: 33579375; PMCID: PMC7881657. - Virtual and In Vitro Antiviral Screening Revive Therapeutic Drugs for COVID-19.
Bocci G, Bradfute SB, Ye C, Garcia MJ, Parvathareddy J, Reichard W, Surendranathan S, Bansal S, Bologa CG, Perkins DJ, Jonsson CB, Sklar LA, Oprea TI. ACS Pharmacol Transl Sci. 2020 Oct 14;3(6):1278-1292. doi: 10.1021/acsptsci.0c00131. PMID: 33330842; PMCID: PMC7571299. - REDIAL-2020: A suite of machine learning models to estimate Anti-SARS- CoV-2 activities.
Govinda KC, Bocci G, Verma S, Hassan M, Holmes J, Yang JJ, Sirimulla S, Oprea TI. ChemRxiv [Preprint]. 2020 Sep 16. doi: 10.26434/chemrxiv.12915779. PMID: 33200119; PMCID: PMC7668752. - TCRD and Pharos 2021: mining the human proteome for disease biology.
Sheils TK, Mathias SL, Kelleher KJ, Siramshetty VB, Nguyen DT, Bologa CG, Jensen LJ, Vidović D, Koleti A, Schürer SC, Waller A, Yang JJ, Holmes J, Bocci G, Southall N, Dharkar P, Mathé E, Simeonov A, Oprea TI. Nucleic Acids Res. 2021 Jan 8;49(D1):D1334-D1346. doi: 10.1093/nar/gkaa993. PMID: 33156327; PMCID: PMC7778974. - DrugCentral 2021 supports drug discovery and repositioning.
Avram S, Bologa CG, Holmes J, Bocci G, Wilson TB, Nguyen DT, Curpan R, Halip L, Bora A, Yang JJ, Knockel J, Sirimulla S, Ursu O, Oprea TI. Nucleic Acids Res. 2021 Jan 8;49(D1):D1160-D1169. doi: 10.1093/nar/gkaa997. PMID: 33151287; PMCID: PMC7779058. - Artificial intelligence, drug repurposing and peer review.
Levin JM, Oprea TI, Davidovich S, Clozel T, Overington JP, Vanhaelen Q, Cantor CR, Bischof E, Zhavoronkov A. Nat Biotechnol. 2020 Oct;38(10):1127-1131. doi: 10.1038/s41587-020-0686-x. PMID: 32929264. - Deciphering the Plasma Proteome of Type 2 Diabetes.
Elhadad MA, Jonasson C, Huth C, Wilson R, Gieger C, Matias P, Grallert H, Graumann J, Gailus-Durner V, Rathmann W, von Toerne C, Hauck SM, Koenig W, Sinner MF, Oprea TI, Suhre K, Thorand B, Hveem K, Peters A, Waldenberger M. Diabetes. 2020 Dec;69(12):2766-2778. doi: 10.2337/db20-0296. Epub 2020 Sep 14. PMID: 32928870; PMCID: PMC7679779. - Off-Patent Drug Repositioning.
Avram S, Curpan R, Halip L, Bora A, Oprea TI. J Chem Inf Model. 2020 Dec 28;60(12):5746-5753. doi: 10.1021/acs.jcim.0c00826. Epub 2020 Sep 15. PMID: 32877182. - QSAR without borders.
Muratov EN, Bajorath J, Sheridan RP, Tetko IV, Filimonov D, Poroikov V, Oprea TI, Baskin II, Varnek A, Roitberg A, Isayev O, Curtarolo S, Fourches D, Cohen Y, Aspuru-Guzik A, Winkler DA, Agrafiotis D, Cherkasov A, Tropsha A. Chem Soc Rev. 2020 Jun 7;49(11):3525-3564. doi: 10.1039/d0cs00098a. Epub 2020 May 1. Erratum in: Chem Soc Rev. 2020 May 22;:. Curtalolo, Stefano [corrected to Curtarolo, Stefano]. PMID: 32356548; PMCID: PMC8008490. - How many rare diseases are there?
Haendel M, Vasilevsky N, Unni D, Bologa C, Harris N, Rehm H, Hamosh A, Baynam G, Groza T, McMurry J, Dawkins H, Rath A, Thaxton C, Bocci G, Joachimiak MP, Köhler S, Robinson PN, Mungall C, Oprea TI. Nat Rev Drug Discov. 2020 Feb;19(2):77-78. doi: 10.1038/d41573-019-00180-y. PMID: 32020066; PMCID: PMC7771654. - Will Artificial Intelligence for Drug Discovery Impact Clinical Pharmacology?
Zhavoronkov A, Vanhaelen Q, Oprea TI. Clin Pharmacol Ther. 2020 Apr;107(4):780-785. doi: 10.1002/cpt.1795. Epub 2020 Mar 3. PMID: 31957003; PMCID: PMC7158211. - How to Illuminate the Druggable Genome Using Pharos.
Sheils T, Mathias SL, Siramshetty VB, Bocci G, Bologa CG, Yang JJ, Waller A, Southall N, Nguyen DT, Oprea TI. Curr Protoc Bioinformatics. 2020 Mar;69(1):e92. doi: 10.1002/cpbi.92. PMID: 31898878; PMCID: PMC7818358. - Can BDDCS illuminate targets in drug design?
Bocci G, Benet LZ, Oprea TI. Drug Discov Today. 2019 Dec;24(12):2299-2306. doi: 10.1016/j.drudis.2019.09.021. Epub 2019 Oct 1. PMID: 31585170; PMCID: PMC7002033. - Exploring the dark genome: implications for precision medicine.
Oprea TI. Mamm Genome. 2019 Aug;30(7-8):192-200. doi: 10.1007/s00335-019-09809-0. Epub 2019 Jul 4. PMID: 31270560; PMCID: PMC6836689. - The human endogenous metabolome as a pharmacology baseline for drug discovery.
Bofill A, Jalencas X, Oprea TI, Mestres J. Drug Discov Today. 2019 Sep;24(9):1806-1820. doi: 10.1016/j.drudis.2019.06.007. Epub 2019 Jun 19. PMID: 31226432; PMCID: PMC7748399. - How to Prepare a Compound Collection Prior to Virtual Screening.
Bologa CG, Ursu O, Oprea TI. Methods Mol Biol. 2019;1939:119-138. doi: 10.1007/978-1-4939-9089-4_7. PMID: 30848459. - How to Develop a Drug Target Ontology: KNowledge Acquisition and Representation Methodology (KNARM).
Küçük McGinty H, Visser U, Schürer S. Methods Mol Biol. 2019;1939:49-69. doi: 10.1007/978-1-4939-9089-4_4. PMID: 30848456; PMCID: PMC7257161. - DrugCentral 2018: an update.
Ursu O, Holmes J, Bologa CG, Yang JJ, Mathias SL, Stathias V, Nguyen DT, Schürer S, Oprea T. Nucleic Acids Res. 2019 Jan 8;47(D1):D963-D970. doi: 10.1093/nar/gky963. PMID: 30371892; PMCID: PMC6323925. - Darkness in the Human Gene and Protein Function Space: Widely Modest or Absent Illumination by the Life Science Literature and the Trend for Fewer Protein Function Discoveries Since 2000.
Sinha S, Eisenhaber B, Jensen LJ, Kalbuaji B, Eisenhaber F. Proteomics. 2018 Nov;18(21-22):e1800093. doi: 10.1002/pmic.201800093. Epub 2018 Oct 30. PMID: 30265449; PMCID: PMC6282819. - Proteomic analysis defines kinase taxonomies specific for subtypes of breast cancer.
Collins KAL, Stuhlmiller TJ, Zawistowski JS, East MP, Pham TT, Hall CR, Goulet DR, Bevill SM, Angus SP, Velarde SH, Sciaky N, Oprea TI, Graves LM, Johnson GL, Gomez SM. Oncotarget. 2018 Jan 29;9(21):15480-15497. doi: 10.18632/oncotarget.24337. PMID: 29643987; PMCID: PMC5884642. - Unexplored therapeutic opportunities in the human genome.
Oprea TI, Bologa CG, Brunak S, Campbell A, Gan GN, Gaulton A, Gomez SM, Guha R, Hersey A, Holmes J, Jadhav A, Jensen LJ, Johnson GL, Karlson A, Leach AR, Ma'ayan A, Malovannaya A, Mani S, Mathias SL, McManus MT, Meehan TF, von Mering C, Muthas D, Nguyen DT, Overington JP, Papadatos G, Qin J, Reich C, Roth BL, Schürer SC, Simeonov A, Sklar LA, Southall N, Tomita S, Tudose I, Ursu O, Vidovic D, Waller A, Westergaard D, Yang JJ, Zahoránszky-Köhalmi G. Nat Rev Drug Discov. 2018 May;17(5):377. doi: 10.1038/nrd.2018.52. Epub 2018 Mar 23. Erratum for: Nat Rev Drug Discov. 2018 May;17 (5):317-332. PMID: 29567993. - Unexplored therapeutic opportunities in the human genome.
Oprea TI, Bologa CG, Brunak S, Campbell A, Gan GN, Gaulton A, Gomez SM, Guha R, Hersey A, Holmes J, Jadhav A, Jensen LJ, Johnson GL, Karlson A, Leach AR, Ma'ayan A, Malovannaya A, Mani S, Mathias SL, McManus MT, Meehan TF, von Mering C, Muthas D, Nguyen DT, Overington JP, Papadatos G, Qin J, Reich C, Roth BL, Schürer SC, Simeonov A, Sklar LA, Southall N, Tomita S, Tudose I, Ursu O, Vidovic D, Waller A, Westergaard D, Yang JJ, Zahoránszky-Köhalmi G. Nat Rev Drug Discov. 2018 May;17(5):317-332. doi: 10.1038/nrd.2018.14. Epub 2018 Mar 23. Erratum in: Nat Rev Drug Discov. 2018 Mar 23;: PMID: 29472638; PMCID: PMC6339563. - Pharos: Collating protein information to shed light on the druggable genome.
Nguyen DT, Mathias S, Bologa C, Brunak S, Fernandez N, Gaulton A, Hersey A, Holmes J, Jensen LJ, Karlsson A, Liu G, Ma'ayan A, Mandava G, Mani S, Mehta S, Overington J, Patel J, Rouillard AD, Schürer S, Sheils T, Simeonov A, Sklar LA, Southall N, Ursu O, Vidovic D, Waller A, Yang J, Jadhav A, Oprea TI, Guha R. Nucleic Acids Res. 2017 Jan 4;45(D1):D995-D1002. doi: 10.1093/nar/gkw1072. Epub 2016 Nov 29. PMID: 27903890, PMCID: PMC5210555 - Unexplored opportunities in the druggable human genome
T. Oprea, et al., Nature Rev. Drug Discov. Poster Jan. 2017.
