
Knowledge Management Center for Illuminating the Druggable Genome

Lars Juhl Jensen, Ph.D.
Professor
Cellular Network Biology
Novo Nordisk Foundation
Center for Protein Research
University of Copenhagen, Denmark
Professor
Cellular Network Biology
Novo Nordisk Foundation
Center for Protein Research
University of Copenhagen, Denmark
Contacts
- Lars Juhl Jensen’s group:https://www.cpr.ku.dk/research/disease-systems-biology/jensen/
- Lars Juhl Jensen’s lab: https://jensenlab.org/
![]() Lars Juhl Jensen: @larsjuhljensen
Lars Juhl Jensen: @larsjuhljensen
![]() LJJ Facebook
LJJ Facebook
Overview
Work by Dr. Lars Juhl Jensen has resulted in improved text-mining technology that is applied to scientific literature for scoring how well studied each target protein may be and to support target prioritization. This text-mining platform also helps provide tissue and disease associations for the targets. Current Post-Doctoral fellow Dr. Alexander Junge contribute to this work.
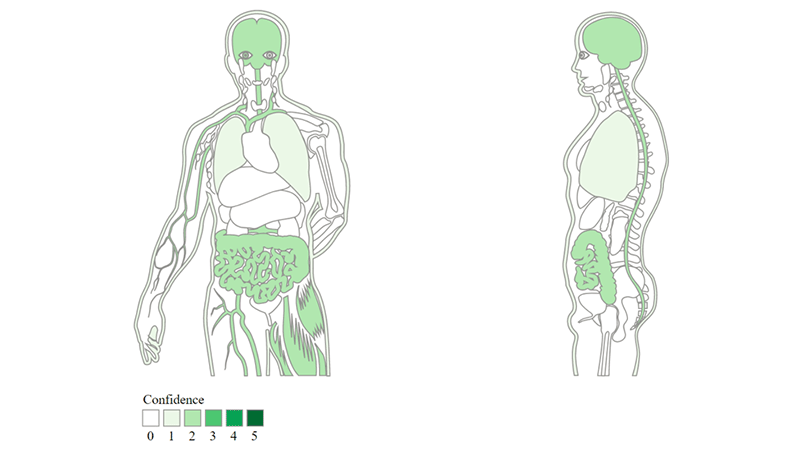
Image depicts tissue expression data extracted by Jensen’s lab via text mining for GPCR, Substance-P receptor (https://pharos.nih.gov/idg/targets/P25103), with the intensity of green displaying the confidence of supporting data for tissue expression.
NIH grant number: 1U24 CA224370-01
KMC-CPR publications:
- Darkness in the Human Gene and Protein Function Space: Widely Modest or Absent Illumination by the Life Science Literature and the Trend for Fewer Protein Function Discoveries Since 2000. Sinha S, Eisenhaber B, Jensen LJ, Kalbuaji B, Eisenhaber F. Proteomics. 2018 Nov;18(21-22):e1800093. doi: 10.1002/pmic.201800093. Epub 2018 Oct 30. PMID: 30265449
- Unexplored therapeutic opportunities in the human genome. Oprea TI, Bologa CG, Brunak S, Campbell A, Gan GN, Gaulton A, Gomez SM, Guha R, Hersey A, Holmes J, Jadhav A, Jensen LJ, Johnson GL, Karlson A, Leach AR, Ma'ayan A, Malovannaya A, Mani S, Mathias SL, McManus MT, Meehan TF, von Mering C, Muthas D, Nguyen DT, Overington JP, Papadatos G, Qin J, Reich C, Roth BL, Schürer SC, Simeonov A, Sklar LA, Southall N, Tomita S, Tudose I, Ursu O, Vidovic D, Waller A, Westergaard D, Yang JJ, Zahoránszky-Köhalmi G. Nat Rev Drug Discov. 2018 May;17(5):317-332. doi: 10.1038/nrd.2018.14. Epub 2018 Mar 23. PMID: 29472638
- Drug target ontology to classify and integrate drug discovery data. Lin Y, Mehta S, Küçük-McGinty H, Turner JP, Vidovic D, Forlin M, Koleti A, Nguyen DT, Jensen LJ, Guha R, Mathias SL, Ursu O, Stathias V, Duan J, Nabizadeh N, Chung C, Mader C, Visser U, Yang JJ, Bologa CG, Oprea TI, Schürer SC. J Biomed Semantics. 2017 Nov 9;8(1):50. doi: 10.1186/s13326-017-0161-x. PMID: 29122012
- TIN-X: target importance and novelty explorer. Cannon DC, Yang JJ, Mathias SL, Ursu O, Mani S, Waller A, Schürer SC, Jensen LJ, Sklar LA, Bologa CG, Oprea TI. Bioinformatics. 2017 Aug 15;33(16):2601-2603. doi: 10.1093/bioinformatics/btx200. PMID: 28398460
