IDG Consortium
IDG Consortium
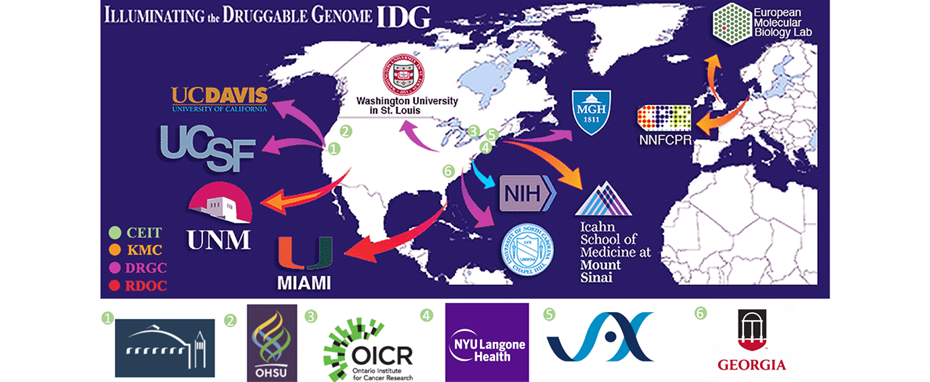
Sponsorship by NIH’s Common Fund has established the program called Illuminating Druggable Genome (IDG) Consortium with the aim of highlighting current knowledge of protein targets through integration of informatics tools, and further study the function of specific understudied targets in three main druggable protein families: G-protein coupled receptors, Ion Channels and protein kinases. This consortium consists of a network of Data and Resource Generation Centers (DRGCs), each focusing their research on one of the three protein families, the Knowledge Management Center (KMC) organizing data and integrated informatics tools across various resources to illuminate understudied protein targets, and the Resource Dissemination and Outreach Center (RDOC) facilitating annotation and distribution of resources brought forth to shed light on to targets. In the spring of 2019, three additional groups joined IDG focusing on making Cutting Edge Informatics Tools for Illuminating the Druggable Genome (CEIT).